Current Projects
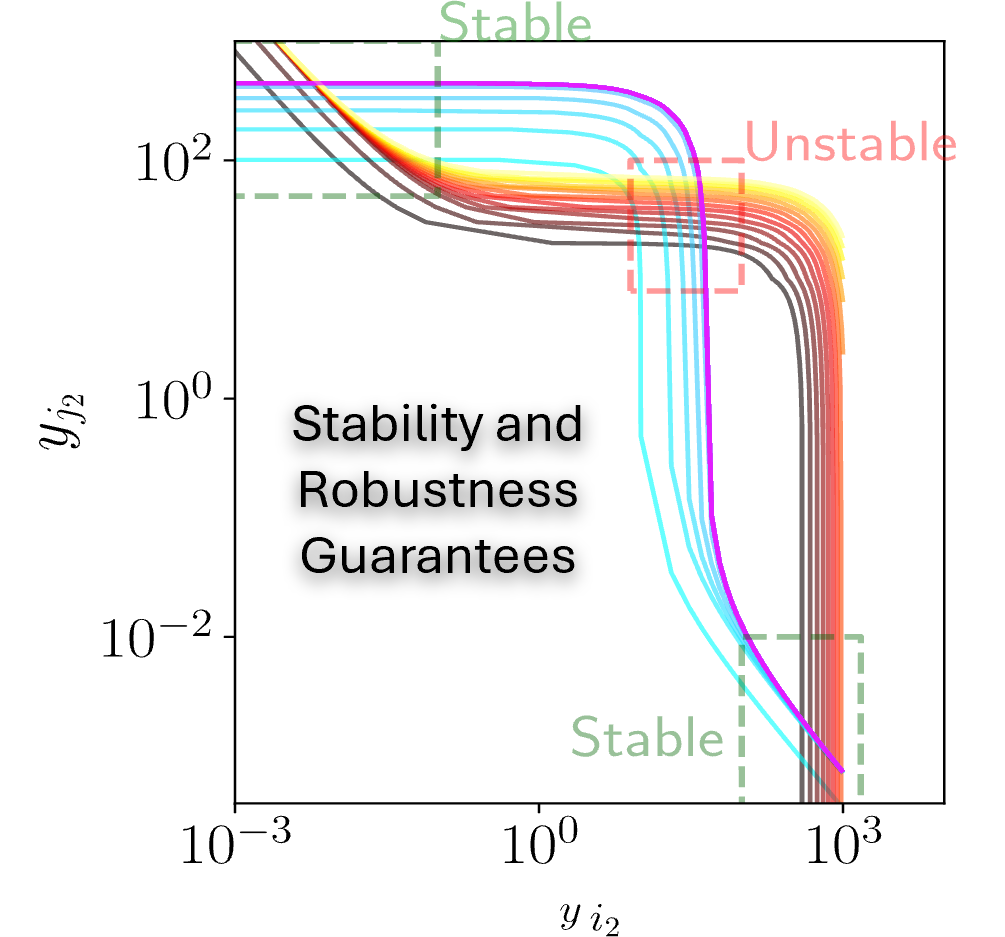
- Parameter robustness in dynamical system models
- Revolutionizing engineering education using collaborative design and integration of sociotechnical modules
- Safety guarantees and robustness quantification of generative AI models
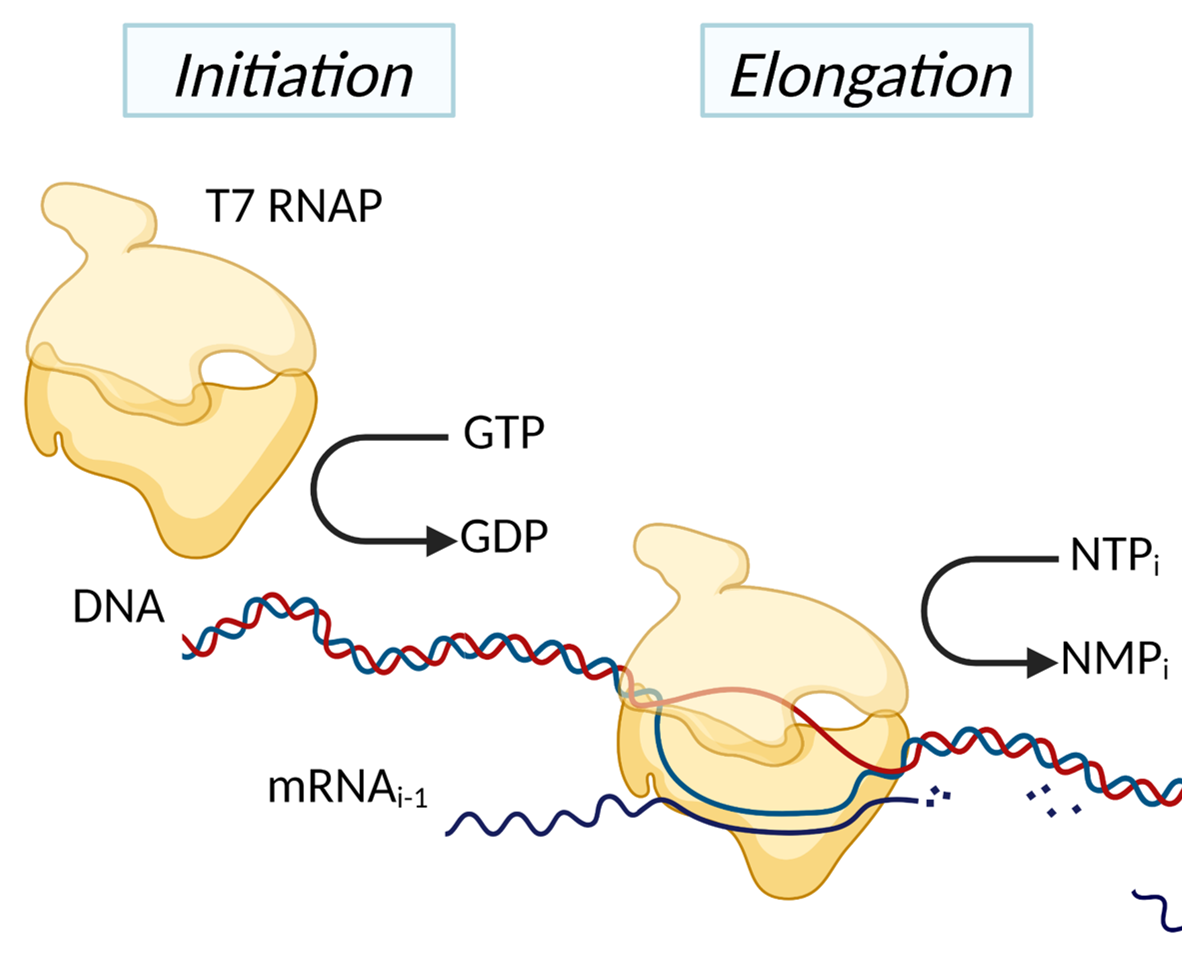
- Robust design and analysis of engineered biological systems
- LLM-based personalized grading to enable independent and open-ended summative assessments
Summer Research:
Available projects for 2026
Involvement in Other Projects and Collaborations
- Mathematical modeling and control of bacterial growth (with Chelsea Hu at Texas A&M University). Read more here.
- Robustness and stability guarantees for nonlinear dynamical systems (with Inigo Incer at University of Michigan; Eduardo Sontag at Northeastern University; Nick Nolan, Emma Peterman, Katie Galloway, and Domitilla Del Vecchio at MIT). Read more here and here.
- Characterization of integrase and excisionase activity in cell-free (with Makena Rodriguez and Richard Murray at Caltech, and William Poole at Altos Labs). Read more in ACS Synthetic Biology.
- Open-source modeling tools for synthetic biology: BioCRNpyler, Bioscrape, Sub-SBML, and AutoReduce. Work in collaboration with William Poole (Altos Labs) and Richard Murray (Caltech)
- Design and implementation of an autonomous bicycle: The i-Bike Project. More here.
Fetching last updated date...
You can contribute to this page by creating a pull request on GitHub. The content of this webpage is licensed under the MIT License.